ATAC with Select Antigen Profiling by sequencing (ASAP-seq) is a method combining sensitive detection of proteins with high quality measurements of chromatin accessibility by single cell ATAC-seq.
ASAP-seq is to scATAC-seq what CITE-seq is to scRNA-seq.
For now, we have demonstrated ASAP-seq on the 10x Genomics scATAC assay, but it should be also compatible with other single cell ATAC methods, e.g. from BioRad.
ASAP-seq makes use of the same reagents used for CITE-seq now available as TotalSeq™ reagents from BioLegend. To make these reagents compatible with scATAC-seq, we use a bridge oligo specific to the appropriate TotalSeq product. With addition of this single bridge oligo, researchers can use their existing CITE-seq or TotalSeq reagents to measure chromatin accessibility together with their favorite proteins. Hashing is also a feature of ASAP-seq, with the same bridge oligo enabling the capture of hashtags. Importantly, ASAP-seq is compatible with sequencing-based detection of intracellular epitopes.
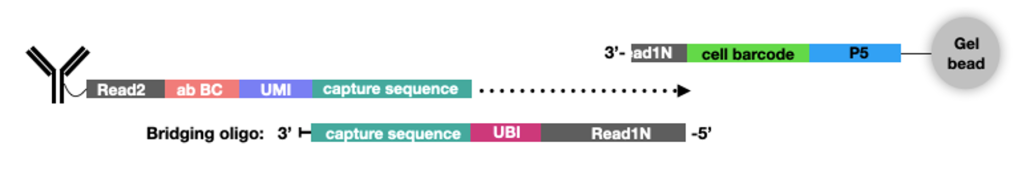
On the sample prep side, ASAP-seq builds on the recently published mtscATAC-seq method, using fixed whole cells as input into scATAC, delivering, in addition to protein information, high quality ATAC-seq data with optional retention of mtDNA for determination of clonal relationships between cells.
ASSAY SCHEMES
ASAP-seq with TotalSeq™-A Hashing reagents
PROTOCOL
Please check out our FAQ or contact us with any questions